2009 Senior Thesis Abstracts
(CLASS OF 2009: If your thesis abstract is not currently included on this page and you would like it to be, please follow this link.)
Inhibiting Secretion during Stimulation Increases Localization of Pro-Egg Laying Hormone mRNA in the Neurites of Bag Cells
Laura Anne Bradley
(Advisor: Arch)
Neuronal activity can stimulate hormone secretion and local synthesis of the hormone precursor. The mRNA coding the prohormone is specifically sent to the site of translation. Previous research shows evidence of stimulation-dependent pro-egg laying hormone (pro-ELH) mRNA trafficking to the neuritic extensions of bag cells of Aplysia californica, located in the pleural visceral connective nerve (PVN). The abdominal ganglion contains two distinct, homogenous populations of bag cells and associated neurites, comprising the bag cell organs (BCOs). In this study it was confirmed that these BCOs have the same response to a high K+-mediated depolarization event. It was further confirmed that depolarization increases pro-ELH transcripts in the PVN. The pro-ELH mRNA was quantified using RT-qPCR, normalized with β-actin transcript levels. Immunohistological experiments on cross-sections from the PVN were performed using two anti-ribosome antibodies and resulted in nonspecific binding. These sections revealed that bag cells extend into the PVN to a greater extent than previously believed. In an attempt to monitor secretion of ELH, the secretion media were stained with a custom anti-ELH antibody. The dot blots demonstrated nonspecific binding of either the primary or secondary, HRP-conjugated antibody and resulted in positive signal in negative controls. The BCO was treated with a low-Ca2+, high-K+ solution that allows depolarization but prevents exocytosis in the bag cells. Inhibiting secretion during depolarization was shown to increase pro-ELH mRNA localization in the bag cell cluster (BCC) and to a greater extent in the PVN. Using BAPTA-AM to inhibit secretion during stimulation resulted in an increase in pro-ELH transcripts in the BCC and a decrease in the PVN, but these data may be confounded by an improper section in the BAPTA-AM treatment BCO and the possibility that BAPTA-AM depolymerized that microtubules and prevented mRNA trafficking. These results suggest that depolarization is the signal for bag cells to traffick pro-ELH mRNA to the neurites. Furthermore, it is possible that in the absence of secretion, translation does not occur and the untranslated mRNAs in the neurites have a longer half-life due to less degradation. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
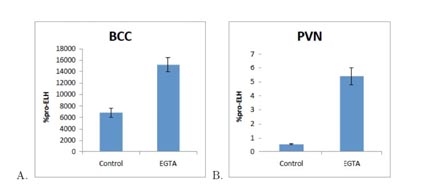
Figure: EGTA-mediated Ca2+ chelation increases pro-ELH mRNA in the PVN and BCC. A. Incubation in EGTA (5µM), low-Ca2+ (1 mM), relative to no EGTA, normal Ca2+ (13 mM) stimulation media increased pro-ELH mRNA in the BCC. B. Incubation in EGTA (5 µM), low-Ca2+ (1mM), relative to no EGTA, normal-Ca2+ (13 mM) stimulation media increased pro-ELH mRNA in the PVN-P. The BCO halves were pretreated in normal or low-Ca2+, basal K+ for 60 minutes, stimulated in normal or low-Ca2+, high-K+ (107 mM) for 60 minutes, post-treated with normal or low-Ca2+, basal-K+ for 60 minutes and allowed to recover in recovery medium for 4 hours before RNA extraction. %pro-ELH represents the quantity of pro-ELH mRNA relative to actin mRNA. (n=3)
Ecological Analysis of Spatial and Temporal Patterns in Distribution and Abundance of Fished in the Reed Canyon Stream
Laila Bryant
(Advisor: Kaplan)
The Reed Canyon is a unique natural habitat that houses the headwaters of Crystal Springs Creek within an urban environment. The Reed Canyon section of the creek and the surrounding riparian area have undergone intensive restoration and management over the past nine years, with the effects of this work going for the most part un-studied during that time. I employed a passive trapping methodology at five sites in the canyon stream, documenting species counts and standard lengths of fish encountered at each site over a five-month period from October 2008 to March 2009 (a total of 30, 72-hour trapping periods). This study was carried out at a finer scale than previous sampling by City and State crews, providing a more detailed and complete picture of assemblage dynamics in the canyon. Using these data, I tested hypotheses pertaining to the temporal and spatial variation in species diversity, abundances, and size of fishes in the Reed Canyon stream assemblage. Greatest diversity was observed at the site furthest downstream, below the SE 28th Ave. culvert, while greatest abundances of fishes were observed at the two most upstream sites, inside the fish ladder. Dramatic seasonal variation in abundances of reticulate sculpin and threespine stickleback was documented, as well as variation in size of sculpin among sites. The dynamics of fish species distribution and fish assemblage observed among sites are likely regulated by a combination of abiotic and biotic factors, including thermal differences among sites, structural differences in habitat, and competitive interactions between species or size classes. These data develop a baseline set of information about the canyon stream fish community prior to the initiation of the second phase of the Reed Canyon Enhancement Strategy. Temporal and spatial variation in assemblage composition among sites has implications for future monitoring goals, revealing sites of high fish activity, and demonstrating the inadequacy of infrequent, general sampling of the stream reach. I conclude by outlining a protocol for implementing an informative, low-maintenance, long-term monitoring plan. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Testing the Role of the Cellular Organizer of a Spider Embryo
Marion Burrill
(Advisor: Black)
The cumulus is a signaling center in spiders known to be crucial for the normal determination of embryonic axes. When the cumulus is surgically deleted or an associated signaling pathway disrupted, the result is abnormal, radialized development. Surgical deletion experiments have not been repeated since their publication in 1952 when they were done using only one species (A. Holm, Zool. Bidrag Uppsala 25: 293). For my thesis research, I removed cumulus cells as well as cells in the adjacent superficial layer in the spiders Latrodectus mactans and L. geometricus. Some abnormal embryos were obtained, resembling those illustrated by Holm. However, most of these embryos were controls in which non-cumulus cells were removed. The conclusion is that cells receiving a putative signal are as important to normal development as the cells emitting the signal. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

One Bad Mother: Functional Genomics of Maternal Aggression in an African Cichlid Fish
Julia Carleton
(Advisor: Renn)
The cichlid fish Astatotilapia burtoni has served as a model system for social behavior for 30 years, yet nearly all of the studies thus far have focused on the plasticity of male dominant and subordinate social phenotypes. However, female behavior has received little attention, often being described in the literature as simple subordination. Recently a novel phenotype of maternal behavior was identified in the first generation of wild-caught A. burtoni females allowed to associate with their offspring. Upon release of fry, wildstock females become aggressive towards conspecifics and take on the appearance of dominant males, while providing care for their fry. Labstock females, inbred for 30 years, display similar aggression but do not care for their fry. The biological basis of this maternal behavior, and how it differs between stocks, is entirely unknown. This thesis used a systems biology approach to investigate the neuroendocrine and genomic contributions to the novel phenotype of maternal aggression in two unique stocks of A. burtoni. Behavioral observations showed that wildstock are more aggressive after release, attacking males in a neighboring tank more often than labstock females. Plasma testosterone levels were not significantly different between the stocks before or after release of fry. cDNA microarrays revealed an incredible difference in gene expression between stocks, and identified a maternal module of gene expression specific to wildstock females after release. This maternal module included genes previously identified in aggressive dominant males and novel genes not associated with any other phenotype. This thesis establishes a robust model for maternal aggression that can be used to study the genetic underpinnings of this complex phenotype. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
Of Granules and Gastropods: Toward the Isolation and Characterization of Neuronal Ribonucleoprotein Particles Containing a Prohormone mRNA
Todd Dembo
(Advisor: Arch)
The localization of mRNA is partially responsible for establishing functional polarity in cells that require tightly regulated control over the spatial and temporal aspects of translation. The unique, reticulated morphology of the neuron necessitates such localization to ensure that the culmination of gene expression occurs in a timely manner. However, the inherent instability of mRNA and apparent lack of translational machinery in neuronal processes raise serious questions regarding how local translation is accomplished. These issues are resolved, in part, by the effectors of mRNA localization: large proteinaceous aggregates known as RNA granules. Actively transported along microtubules, these massive ribonucleoprotein particles are, on average, 200 nm in diameter, and capable of storing up to thirty mRNA transcripts. Isolated granules have been observed to contain a variety of protein components required for RNA transport and protein synthesis. Interestingly, the bag cell neurites of the California sea hare, Aplysia californica, have been shown to contain mRNA encoding for pro-egg laying hormone (pro-ELH), and might be capable of locally synthesizing and secreting both pro- and mature ELH peptides. Evidence also suggests that ELH mRNA is routed to the neurites in an active transport, rather than diffusive manner. Herein, we test the hypothesis that some neuronal granules from the Aplysia abdominal ganglion contain pro-ELH mRNA. Our initial approach was to develop a procedure for granule isolation, and to follow this isolation with an analysis of the isolated products. However, the isolation procedure appeared to lack sufficient specificity, and will require modification for it to be efficacious. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
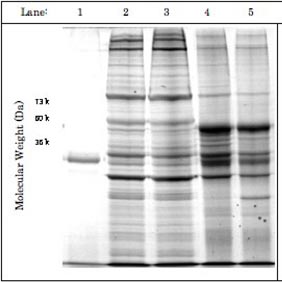
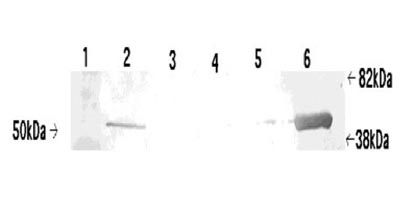
Figure: The products of EF-1α1 immuno-precipitation exhibit some degree of RNase-sensitivity. Two pedal-plural ganglia were homogenized and their supernatant solutions pooled to equalize protein concentrations between samples. Separation by SDS-PAGE (1 = EF-1α1 antibody alone; 2 = Supernatant solution used for immunoprecipitation with RNase inhibitors; 3 = Supernatant solution used for immuno-precipitation with RNase; 4 = RNase inhibitor immunoprecipitation product; 5 = RNase immuno-precipitation product) suggested that RNase treatment exhibited some effect on the efficacy of immunoprecipitation. However, this effect was primarily visible between 73 and 40 kDa, rather than over a broad range of weights. A band present at around 35 kDa was of considerably greater intensity in the RNase immuno-precipitation, though its identity is still unknown.
Visualization of Enterotoxigenic E. coli in the C. elegans Intesetine by Immunofluorence
Dan Glaser
(Advisor: Mellies)
Enterotoxigenic Escherichia coli (ETEC) is a major cause of human infectious diarrhea in developing countries worldwide. ETEC is spread by consumption of food or water contaminated with fecal matter. Once ingested, ETEC adheres to the epithelium of the small intestine, a process mediated by fibrous antigens and short surface proteins. After adhesion, ETEC usually produces two sets of toxins, heat-labile and heat-stable, that induce fluid secretion in the human small intestine, leading to diarrhea. Although it is not traditionally considered in an invasive pathogen, ETEC has the potential to invade intestinal cells. The prototypical ETEC strain H10407 codes for two invasins, TibA and Tia; expression of either protein induces an in vitro invasive phenotype. Though previously validated as a model for other bacteria, the nematode Caenorhabditis elegans has only recently been adapted as a versatile small animal model for ETEC pathogenesis. Kaplan-Meier survival analysis shows that H10407 grown on NGM agar kills nematodes at a significantly faster rate than wild-type E. coli MG1655. H10407 was also shown to colonize the nematode intestine with significantly higher numbers of bacteria than MG1655; fluorescent imaging of infected worms confirmed that large numbers of viable H10407, but not MG1655, are present in the nematode gut. Treatment of infected nematodes with either gentamicin or chloramphenicol significantly reduced intestinal populations of MG1655 but not H10407. Invasion of the nematode's intestinal epithelium was considered as a potential source of protection from chemical treatment. Transmission electron micrographs did not reveal any invasion or disruption of the C. elegans intestinal epithelium. This suggests that ETEC might not invade the intestinal cells in C. elegans, but is nonetheless protected from chemical treatment. Further research into the initial colonization process of ETEC is necessary before any conclusions can be drawn. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
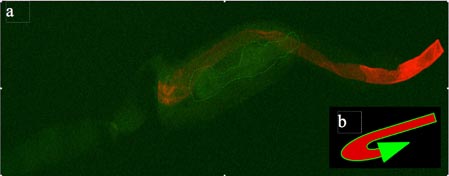
Figure: Fluorescent imaging shows C. elegans intermediate filament web, but no visible ETEC strain H10407
Age synchronized nematodes were incubated overnight on NGM agar plates with pregrown food lawns of either H10407(pKH91) or MG1655(pKH91). Nematodes were then washed and cut randomly before being fixed in formaldehyde. Fixed worms were then freeze-cracked before staining with 1˚ and 2˚ antibodies. Stained worms were visualized by dual-channel confocal fluorescence microscopy. (a) Red corresponds to Alexa-647 stained intestinal web; green is autofluorescenece produced by the nematode carcass. The bilobed grinder visible in the center (dotted line) implies that this is the anterior portion of the nematode. (b) This schematic models the orientation of the nematode anatomy visible in the image itself. Coloration corresponds to those in the image. Scale bars were unavailable in the imaging program.
Isolation of Dense-core Vesicles Toward Identification of Neural Vesicle-associated Proteins
Ben Greenberg
(Advisor: Arch)
Many neurotransmitters and neurohormones are released from neurons by exocytosis from dense-core vesicles (DCVs). To release their contents from axon terminals, DCVs must be transported from the cell body, docked at the membrane and primed for release. Many cytosolic proteins have been identified as factors in each of these steps, but many more remain undiscovered. The bag cells of the marine mollusc Aplysia californica are an excellent system for studying vesicle transport due to their long axons and homogenous population of large dense-core vesicles. I set out to make a qualitative comparison between newly synthesized vesicles in bag cell bodies and vesicles undergoing transport in neurites to identify candidates for novel transport-related proteins. However, a major limitation in the study of vesicleassociated proteins is the lack of a technique for purifying vesicles while maintaining membrane integrity and protein-protein interactions. Here I describe an attempt to develop a technique for immunoprecipitation of whole vesicles and their associated proteins. Immunoprecipitation was performed on tissue homogenates using antibodies to vesicle-associated membrane protein (VAMP) bound to protein A-conjugated paramagnetic beads. This procedure isolated a wide range of proteins, but due to a significant amount of nonspecific binding to the beads, successful isolation of whole vesicles could not be determined. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Is the Interaction Between Human Telomere Proteins TIN2 and TRF1 Conserved in Xenopus laevis?
Jennifer Jin
(Advisor: Shampay)
Telomeres are the ends of chromosomes that, in humans, contain TTAGGG-repeat DNA sequences bound by protein complexes. By controlling the access of telomeres to telomerase, a reverse transcriptase that adds telomeric DNA repeats to chromosome ends, the protein complexes regulate telomere length. Telomere length maintenance is essential for prolonged cell proliferation by protecting the ends from degradation or fusion. In humans, TIN2 (TRF1-Interacting Nuclear protein 2) modulates activity of TRF1 (Telomeric Repeat binding Factor 1) at the telomere, and this activity includes length regulation in the presence of telomerase. Homologous TRF1 and TIN2 genes have been found in Xenopus laevis: xlTRF1 and xlTIN2. Xenopus telomeres also have the TTAGGG repeats, but telomerase is active in both somatic and gamete cells, whereas in humans it is generally repressed in somatic cells. Whether the Xenopus telomere proteins interact was tested by creating His-tagged xlTIN2 and Flag-tagged xlTRF1 and expressing them in vitro. The proteins were successfully immunoprecipitated from rest of the in vitro reactions by antibodies against the corresponding epitope tags. However, the protein-protein interaction by co-immunoprecipitation could not be evaluated due to non-specific immunoprecipitation of each epitope-tagged protein by both antibodies. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program and a grant to JS from the National Science Foundation)
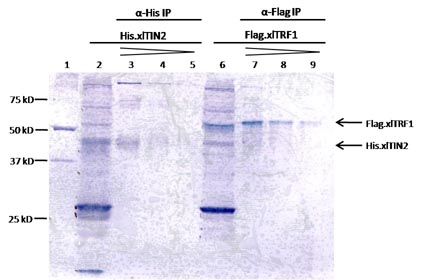
Figure: Immunoprecipitated xlTIN2 and xlTRF1.
His.xlTIN2 and Flag.xlTRF1 proteins were generated separately in vitro in 50 µL reticulocyte lysate reactions. Translated products were immunoprecipitated from 45 µL of each reaction: His.xlTIN2 with anti-His antibody and Flag.xlTRF1 with anti-Flag antibody. Proteins were eluted in 1X SDS sample buffer, and 50%, 25% and 12.5% of eluate were analyzed by 10% SDS-PAGE (lanes 3-6 and 7-9). Ten percent input of each in vitro translation reactions were also analyzed (lanes 2 and 6). Five microliter of unstained marker was used as a reference for molecular weight (lane 1).
Classification of Indoleamine-2,3-dioxygenase in Xenopus laevis as Compare to Humans
Candice Hoi-yan Kam
(Advisor: Reuben)
Indoleamine 2,3-dioxygenase (IDO) is an enzyme that catabolizes L-tryptophan and forms kynurenines as byproducts. In effect, the combination of tryptophan depletion and the presence of kynurenines cause inhibition of thymus derived lymphocytes (T cells). This inhibition creates an immune tolerant microenvironment. Under certain circumstances, such as pregnancy and autoimmune disease, IDO activity serves to protect against the harmful activity of T cells. However, when IDO is over-expressed in other situations, it can be harmful, as in the case of cancer development. There are two different types of IDO: IDO1 and IDO2. These two enzymes can be distinguished through differences in efficiency in tryptophan degradation, with IDO 1 being a much higher efficiency enzyme than IDO 2. It is important from an evolutionary perspective, to determine which type of IDO is active in Xenopus laevis, and because of two unique characteristics of Xenopus. One interesting trait is its resistance to cancer, and the other is immune tolerance during metamorphosis, where the larval immune system fails to reject antigenically disparate adult cells as they differentiate. It has been suggested by others that IDO 2 is the only type of IDO present in Xenopus laevis. A low efficiency IDO might be at least partially responsible for successful immune rejection of antigenically distinct cancer cells in Xenopus. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
Production of Polyhydroxybutyrate in Euphorbia pulcherrima
Hyeon Jeong Kim
(Advisor: Dalton)
The accumulation of non-biodegradable plastics has become a serious environmental problem. Polyhydroxybutyrate (PHB) is a naturally occurring polymer, which has similar characteristics to conventional plastic. PHB, within a year, is fully biodegradable by microorganisms into carbon dioxide and water and therefore has been studied as an alternative to conventional plastic. The production of PHB has been attempted in various plants. However, the diversion of an essential compound, acetyl-CoA, from metabolic pathways to PHB production leads to low PHB accumulation and growth retardation in transgenic plants. Poinsettia, Euphorbia pulcherrima produces a milky sap called latex, which is synthesized from acetyl-CoA. By using this pool of acetyl-CoA for the production of PHB, the detrimental effects to the plants are expected to be minimized while also producing a higher level of PHB. Poinsettia were transformed by Agrobacterium tumefaciens to contain all the genes necessary for PHB production. As of writing, transgenic calli have produced somatic embryos and shoot and root growth are being induced. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

An Investigation into the Interaction of X. laevis telomeric Proteins TRF1 and PinX1
Molly King
(Advisor: Shampay)
The chromosome replication process causes the ends of chromosomes to slowly degrade without the remediative action of the enzyme telomerase. Telomeres are repetitive sequences at the ends of chromosomes that protect them from inappropriate DNA repair and cellular responses to DNA damage. Telomerase and other telomere-associated proteins are essential for end protection, chromosome maintenance and telomere regeneration. Two telomere protection complex proteins, TRF1 and PinX1, have been found to interact and to negatively regulate telomere length in humans. Human PinX1 is a direct inhibitor of telomerase enzymatic activity, while TRF1 indirectly regulates telomerase by preventing access to the telomeric overhang. Xenopus laevis has previously been shown to have constitutive telomerase activity in somatic cells, making it an excellent model organism for research on telomerase and telomere length regulation. This study sought to investigate the interaction of the telomeric proteins TRF1 and PinX1 in Xenopus laevis. An in vitro expression construct for a Myc-epitope tagged TRF1 dimerization domain was created to aid in the study of this interaction. This construct can be used in future studies for more specific identification of interacting domains, should PinX1 and full-length TRF1 be found to interact in X. laevis. The interaction of X. laevis PinX1 and full-length TRF1 was investigated using previously cloned constructs. Immunoprecipitation and coimmunoprecipitation studies were performed to investigate in vitro interaction. While no conclusive evidence was found either for or against interaction, progress was made towards a rigorous experimental design to test this interaction. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program and a grant to JS from the National Science Foundation)
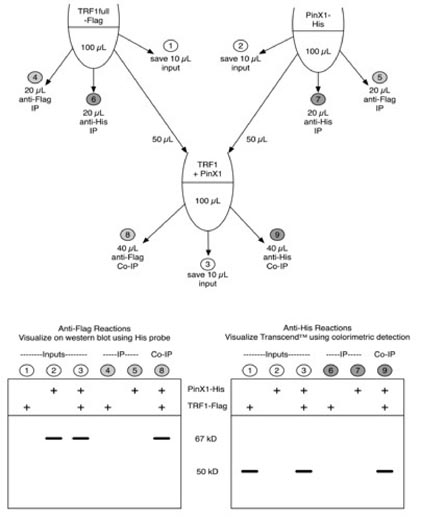
Figure: Schema of experimental design for immunoprecipitation (IP) and co-IP reactions. Top two "tubes" represent inputs of in vitro translated xlTRF1full-Flag and E. coli expressed xlPinX1-His (mixed with reticulocyte transcription/translation reagents as described in the methods) proteins. These reactions were split in half, one half for IPs and one half for protein interaction experiments. Lower "tube" represents incubation of PinX1 and TRF1 together, which then was split for two co-IPs, one each using antibodies to Flag or His. Light grey circles correspond to lane numbers visualized on western blot using His probe. Dark grey circles correspond to lane numbers visualized using colorimetric detection of biotin labeled amino acids. White circles represent lanes included in both analyses.
The Putative Testis-differentiating Gene, Dmrt1, is Expressed in Both Males and Females During Sex-determination in Threespine Stickleback (Gasterosteus aculeatus)
Jordan Kohn
(Advisor: McClellan)
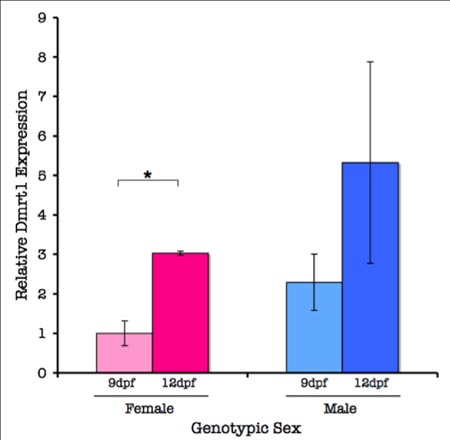
The differentiation of organisms into male versus female sexes is both a crucial developmental event and a necessary precondition for sexual reproduction. Interestingly, no universal mechanism for this process has been discovered. In the threespine stickleback (Gasterosteus aculeatus ), male gonadal development is influenced by an unidentified sex-determining factor (SDF ) on the Y chromosome, which inhibits primordial germ cell (PGC) proliferation. A testis-differentiating gene, Dmrt1, may regulate this process in stickleback. In this investigation, I used molecular markers to assign genetic sex and real-time quantitative polymerase chain reaction (RT-qPCR) to analyze Dmrt1 expression in stickleback juveniles during the sex-determining phase of development. I also used a PGC-specific riboprobe to track PGC migration to the gonad. I found a trend toward a two-fold increase in Dmrt1 expression during PGC colonization in males versus females. Attempts to visualize PGC migration during colonization using a DIG-labeled riboprobe were unsuccessful. These findings indicate that Dmrt1 may be important as an immediate downstream mediator or coregulator of male development in threespine stickleback. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
Glucosinolates and Selenium Might Interact in the Face of Adversity to Form an Anti-Herbivore Superpower Coalition
Lillian Kuehl
(Advisor: Karoly)
Selenium is a mineral required for animal, but not plant, health. However, certain plants, dubbed accumulators or hyperaccumulators, take up a disproportionate amount of selenium from the soil into their tissues. Previous research has hypothesized that this is a defensive mechanism, decreasing the amount of herbivory a plant is subjected to. Another way that certain plants, including Indian mustard (Brassica juncea) and other members of Brassicaceae, limit herbivory is the production of toxic organic compounds. Mustards produce glucosinolates, which react when tissue is damaged to produce toxic and bitter-tasting isothiocyanates. Previous research has shown that the production of glucosinolates may be induced by herbivory or wounding. I was interested in whether selenium, as another defensive compound, might be induced in a similar way. I investigate the induction of selenium uptake in plants subjected to artificial wounding, and attempted to analyze the potential interaction with glucosinolate induction. Before high mortality rates forced me to revert to mechanical wounding, I attempted to use white cabbage butterfly (Pieris rapae) larvae as the herbivore treatment. Because selenium and sulfur are chemically similar, and sulfur is an integral part of glucosinolates, I included both a sulfur and a selenium soil treatment. I found strong effects of sulfur on plant growth, and some effect of selenium and sulfur-selenium interaction on plant growth. My results were limited by the failure of my glucosinolate assay, and a small sample size used for selenium analysis. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Introduction of Several Population of a Species of the Endangered Wildflower Delphinium leucophaeum into Two Different Parks in the Northern Williamette Valley, Oregon
Jenny Leonard
(Advisor: Karoly)
Introductions of endangered plants are implemented frequently, but often with little scientific basis. In this thesis, I have explored several introduction variables recommended by the scientific community. Variables include genetics, habitat suitability, and the viability of different types of propagules. The importance of these variables may change from species to species, but they are important to consider while designing and implementing an introduction.
I introduced populations of D. leucophaeum, an endangered wild flower found in Oregon and Washington,to protected sites in the greater Portland area. The experimental goals of this project were to answer questions about the effect of environmental variables on D. leucophaeum. Experiments were used to gather information about the variables that could affect the success of the planting of D. leucophaeum.
I weighed and planted 432 tubers at Canemah Bluffs and 144 at the Camassia Preserve. Tubers were planted in closed canopy sites (Closed), partial shade sites (Partial), and open sites (Open), with 48 tubers at each of 12 sites. There were three replicates of each microhabitat type at Canemah and one replicate at Camassia. Once plants emerged we recorded temperature and light levels at the site using datalogging devices and measured soil moisture. Two censuses were conducted, one in mid-March and the other in mid-April. We recorded width of the largest leaf as a measure of fitness for each individual. Site suitability and quality was determined by the average leaf size at each site. The effects of site, location, tuber size and abiotic factors on leaf size were analyzed.
This project was biologically successful in establishing populations of D. leucophaeum at the Camassia Preserve and Canemah Bluffs. Of the tubers planted that could be later censused, 97% exhibited signs of above-ground growth over the course of the experiment. Of the emerged plants from the first census, 16% disappeared by the second census. We found that plants that disappeared had smaller tubers than plants that survived, and that larger tubers produced larger leaves. We also found that sites with lower average and maximum temperature had larger leaves on average. Closed canopy sites had larger leaves than Partial sites, which in turn had larger leaves than Open sites. These results suggest that plants do better in shady, cool sites. Further monitoring will need to be conducted to see if the plants survive to flower and reproduce. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Aggression in Lab Stock and Wild Stock Male Astatotilapia burtoni
Helen Magee
(Advisor: Renn)
The cichlid species Astatotilapia burtoni has been used as a model system to study social behavior because of its complex social system in which social interactions influence status, behavior and androgen levels. There are two male phenotypes, dominant and subordinate. Males of dominant status will behave aggressively and have higher levels of androgen hormones, such as testosterone, than males of subordinate status. While lab-maintained model system are helpful in gaining an understanding of the mechanisms of biological systems, being reared in a laboratory environment over an extended period of time can result in artificial selection that can have significant behavioral and physiological influences on the species being studied. For example, domestication can occur, which results in reduced aggressive behaviors.
In this thesis, the behavior and androgen levels were compared between two stocks of A. burtoni, the lab stock that has been studied in the laboratory environment for 30 to 50 generation, and the first generation of the wild stock, collected from Lake Tanganyika in 2005. The focus of this study is on aggressive behaviors and androgen hormones in males, with the expectation that the lab stock males would show less aggression and have lower levels of testosterone than the wild stock males.
Results of this experiment were mixed. The aggressive behavior chase occurred more frequently in wild stock males, while the aggressive behaviors side threat and border fight occurred more in the lab stock males. The wild stock males had higher levels of testosterone than the lab stock males.
This study also used microarray analysis to compare gene expression between wild caught dominant and subordinate males. Aromatase, melanin and isomerase were upregulated in the dominant males, and somatotropin, topoisomerase, prolactin and actin were upregulated in the subordinate males. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Astatotilapia burtoni Maternal Mouth-brooding Cichlids as a New Model for Appetite Regulation
My Linh Nguyen
(Advisor: Renn)
Mrowka (1984) found female mouthbrooding cichlids develop partial anorexia during brooding, which prevents them from eating their own fry. That study suggests partial anorexia is controlled by long-term regulation of food intake. By looking at the melanocortin system, this thesis will explore the molecular basis of partial anorexia that is exhibited in female African cichlid, Astatotilapia burtoni during the process of mouthbrooding.
For the first time in A. burtoni, this study cloned and identified agouti-related peptide (AGRP), an orexigenic neuropeptide in the melanocortin system. AGRP is expressed in the form of mRNA in the brain tissues of A. burtoni. The predicted 99-amino acid sequence of AGRP is similar to pufferfish AGRP, the variant that is primarily found in the hypothalamus and pituitary of the brain. This study compared the effects of different feeding conditions on the mRNA expression of melanocortin peptides that are classically known to play crucial roles in long-term feeding regulation. The orexigenic peptides of interest are AGRP and neuropeptide Y (NPY). Female A. burtoni from laboratory stock (LS) and wild stock (WS) experienced three feeding conditions: starved, fed, and brooding.
Though feeding conditions did affect the percent weight changes and the gonad-somatic indexes (GSI) in both fish stocks, real-time quantitative polymerase chain reaction (RT-qPCR) measurements of transcript abundance showed that the feeding conditions did not affect expression of AGRP and NPY mRNA. Because many studies have demonstrated the upregulation of orexigenic AGRP and NPY under starving conditions in other fish species, the similarity under the starved condition to the fed condition in this project suggests the results are inconclusive, and will require further investigation and modifications in the experimental design. By understanding the feeding regulation of self-starvation during mouthbrooding in A. burtoni, one can potentially understand the anorexic nature of the debilitating medical condition of cachexia. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Figure: Multiple sequence alignment of AGRP protein sequences of selected species.
Alignment of predicted amino acid sequences of AGRP in A. burtoni with identified zebrafish (XP_001923939, XP_001334946), goldfish (CAD88212), pufferfish or Torafugu (BAF37115, BAF37116), chicken (BAA82256), mouse (AAH79902), human (NP_001129), pig (NP_001011693), and cow (NP_776408) AGRP protein sequences. Identical amino acids are in white letters and black boxed. Gray boxed letter is conserved substitutions. Dashes were introduced to improve the alignment. The lines that join the cysteine residues indicate the disulfide bonds. Alignment was performed in ClustalW.
Transcriptional Regulation of the Tumor Suppressor Protein p53 and the Proto-Oncogene Mdm2 in Xenopus laevis
Jonathan Packer
(Advisor: Reuben)
The South African Clawed Frog, Xenopus laevis, exhibits high resistance to both spontaneous and induced tumor formation, making it a potentially invaluableanimal model for probing the evolution of cancer resistance in vertebrates. The tumor suppressor protein p53 has been the subject of intense scrutiny for the past 20 years. It regulates cell-cycle arrest or apoptosis in the face of potentially tumorigenic stimuli such as DNA damage and cellular stress. The gene encoding the p53 protein is mutated in over 50% of all human cancer cells. p53 expression is regulated at both the mRNA and protein levels, rising in concentration after oncogenic stimuli. Levels of the p53 protein are negatively regulated by the proto-oncogene Mdm2, an effect that is mitigated during activation of p53. Transcription of the p53 encoding gene region is also upregulated by genotoxic stimuli, while transcription of Mdm2 is downregulated by strong genotoxic stimuli. When active, mammalian p53 is transported from the cytoplasm into the nucleus, where it regulates the transcription of genes involved in cell-cycle progression and apoptosis, resulting in either cell-cycle arrest or cell death. An ortholog to mammalian p53 has been identified in X. laevis (Xp53), exhibiting high evolutionary conservation of both biochemical properties and cellular functions to those of mammalian p53. Similarly, an X. laevis ortholog to mammalian Mdm2 has been characterized. Previous thesis work from this lab suggests that Xp53 is constitutively expressed and not inducible by genotoxic stress (UV irradiation) in adult X. laevis splenocytes. In this thesis, the abundance of Xp53 and Xdm2 transcripts as observed in UV-B irradiated or non-irradiated adult X. laevis splenocytes. Two different Xdm2 paralogs were expressed at different levels in both control and UV treated cells, contrary to published results probing the same system. qPCR was unable to detect a significant UV treatment effect on the regulation of either Xp53 or Xdm2, although the experimental UV dose may have been too large. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
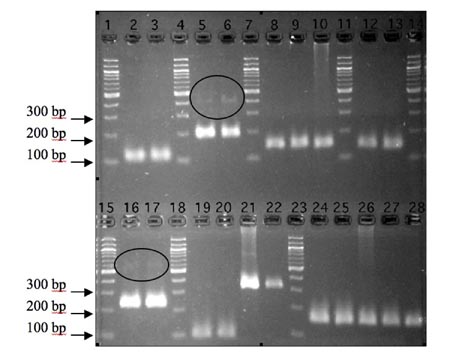
Figure: Primers designed for Xp53, Xdm2_24.1 and GAPDH are cDNA-specific, while those designed for Xdm2_23.1, Ppib1, and COPS3 readily amplify genomic DNA.
Target regions were amplified from either cDNA from control and UV treated splenocytes (left to right), genomic DNA, or no-RT template. Lanes 1, 4, 7, 11, 14, 15, 18, and 23 all contain 100 bp ladder. Lanes 2 & 3 contain the Xp53 target region amplified from cDNA. Lanes 5 & 6 contain the Xdm2_24.1 target region amplified from cDNA. The faint bands around 400 bp could be due to overflowed DNA ladder or amplification of off target sequences. Lanes 8 & 9 contain the Xdm2_23.1 target region amplified from cDNA while lane 10 contains amplified genomic DNA template, resulting in a band exactly the same size as those observed in lanes 8 & 9. Lanes 12 & 13 contain the target Xdm2_23.1 target region amplified from no-RT template, Lanes 16 & 17 contain the X. laevis GAPDH ortholog target region amplified from cDNA. The faint bands at ~500 bp could be due to amplification of a larger, off-target transcript. Lanes 19 & 20 contain the target region of the X. laevis Ppib1 ortholog from cDNA. In lanes 21 & 22, the Ppib1 primers appear to have amplified a ~300 bp target region from genomic DNA and no-RT template, respectively. The discrepancy in size between Ppib1 amplicons from either cDNA, genomic, or no-RT template suggests that, although the primers used are capable of amplifying genomic template, no such amplification occurred when cDNA template was predominant. In lanes 24-28, primers for the X. laevis COPS3 ortholog amplify the same sized target region from cDNA, genomic, and no-RT template. Some bands appear smeared, probably due to both gel overloading and the extended period over which the gel was run (~3 hours). Smears observed in wells 10, 21, and 26 are likely due to the presence of genomic DNA in these particular reactions.
The Role of Soybean Biotin Production on Poly-β-hydroxybutyrate Degradation and Root Nodule Colonization
Elana Peach-Fine
(Advisor: Dalton)
Biological nitrogen fixation accounts for about 60% of the annual global input of fixed nitrogen. Much of this nitrogen is fixed by rhizobia, a group of bacteria that fix nitrogen in symbiosis with legumes. Many biological and environmental factors affect the development of this symbiosis. This project examines the role that nitrogen limitation plays on rhizobial root colonization through its affect on soybean production of biotin. The hypothesized mechanism by which biotin affects rhizobial root colonization is as an upregulator of poly-ß-hydroxybutyrate (PHB) degradation, which in turn could enhance the ability of rhizobia to colonize roots through its use as an internal carbon source. Spectrophotometry was used to measure biotin content of soils in which soybeans had been grown under nitrogen-limiting or nitrogen-sufficient conditions. The hypothesis that biotin affects PHB content was tested in the rhizobia species Sinorhizobium fredii by PHB extraction from these samples and analysis by gas chromatography. Nitrogen limitation resulted in an upregulation of biotin production, but improved methodology could increase confidence in this result. It was also found that PHB degradation was upregulated by the presence of biotin in the bacterial culture medium. These results lend support to the theory that under conditions of nitrogen stress, plant-exuded biotin could act to upregulate the degradation of PHB as a carbon source to competitively aid rhizobia in the formation of nitrogen-fixing nodules on legume roots. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)

Figure: Soybeans at the time of removal from greenhouse. The top photograph shows the secondary planting removed from the greenhouse at 26 days. The bottom photograph shows the tertiary planting removed from the greenhouse at 28 days. In both photographs the nitrogen-stressed plants are the four shown on the left.
Gene Expression Hormones and Behavior in a Sex-role Conventional and Sex-role Reversed Cichlid Species Pair
Molly Schumer
(Advisor: Renn)
This study investigated the relationship between gene expression, hormones, and behavior in a sex-role conventional and sex-role reversed cichlid species pair in the genus Julidochromis. In almost all cichlids behavior is clearly sexually dimorphic. Males are larger and show territorial and aggressive behavior, while females are non-territorial and perform most parental care. Behavioral observations of J. transcriptus have shown that this species conforms to the ancestral pattern in terms of behavioral sexual dimorphism. However, its sister species, J. marlieri, shows an inversion of sex-typical behavior patterns. J. malieri females are larger than males and are clearly the aggressive sex. To examine the changes that accompany a major, heritable change in behavior and mating strategy, I used cDNA microarrays, ELISAs and focal observations to compare J. transcriptus and J. marlieri. Behavioral observations provided compelling evidence for a sex-role conventional phenotype in J. transcriptus and a sex role reversed phenotype in J. marlieri. Analysis of microarray data showed that there are certain gene expression patterns associated with sex –role independent of gonadal sex, and other gene expression patterns associated with sex independent of sex-role. In addition, much of the variation in gene expression observed was species-specific. These data, in combination with comparisons to a model cichlid, suggest that J. marlieri males and females have undergone changes in gene expression patterns leading to a novel behavioral phenotype. This study also contributes to a rich literature on the role of gene expression and hormones in regulating behaviors such as aggression and parental care. (This study was funded in part by a grant from the Reed College Biology Undergraduate Research Program)
Predicting the Potential Distribution of the Invasive Species Brachypodium sylvaticum According to the Niche Conservatism Theory
Angeline Wolski
(Advisor: Karoly)
Niche conservatism theory has taken a forefront in both current biological debate and biological research in the past decade. The theory states that a species will conserve its niche through evolutionary time. Empirical evidence for niche conservatism has been observed through the use of species distribution modeling programs. Programs that model niche conservatism can also predict species distribution in novel locations, such as predicting the spread of invasive species in a new range. This thesis seeks to apply the theory of niche conservatism to predict the potential distribution of the invasive species, false brome, Brachypodium sylvaticum, across Oregon and the Pacific Northwestern United States. Through the use of the Genetic Algorithm for Rule-set Production (GARP), I predicted the potential distribution of B. sylvaticum based on its ecological niche in its native range in Europe. Combinations of a 16,000 point native range occurrence data set and 300 point invasive range occurrence data set were run with 19 bioclimatic environmental layers. Species potential distribution was predicted in the native as well as invaded range. Species distribution was also predicted based on current invasions, unrasterized and rasterized data points, and genetic diversity research. The invaded range modeling predictions diverged from expected niche conserved patterns, providing support for the theory of niche evolution and local adaptation in B. sylvaticum in Oregon. Niche conservatism cannot accurately predict the potential range of some invasive species; therefore, niche evolution may be a factor in allowing for species spread. Management systems should recognize the possibility of niche evolution and local adaptation on invasive species in their management practices.
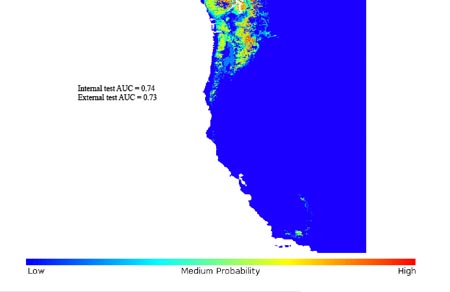
Figure legend to image: Figure 1. Invasive range prediction based on native occurrence points
Potential distribution of B. sylvaticum in its invaded range in the Pacific Northwest of the United States, based on EU_cut occurrence data from its native range in Europe. Trial run with GARP Best Subsets.