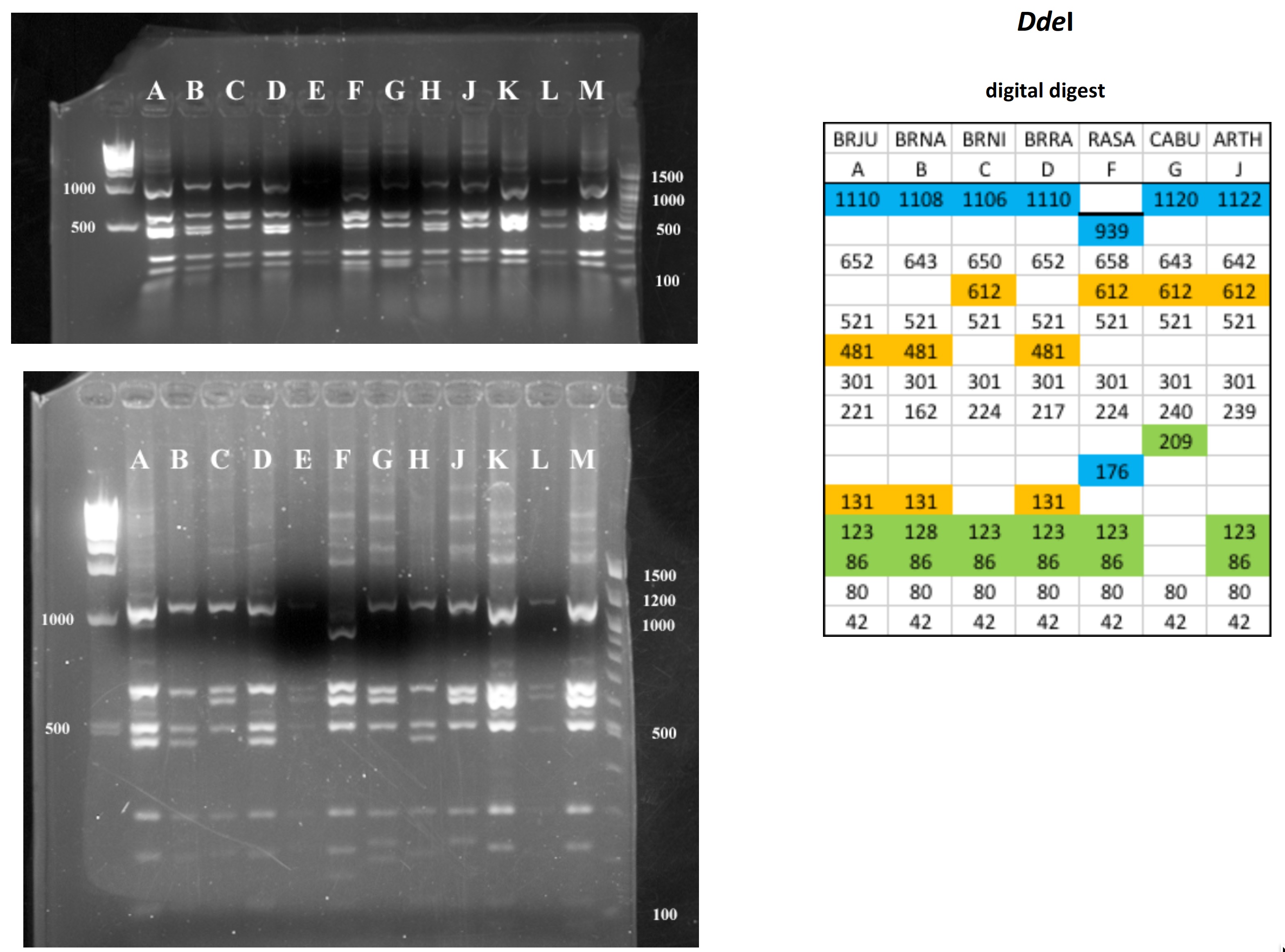
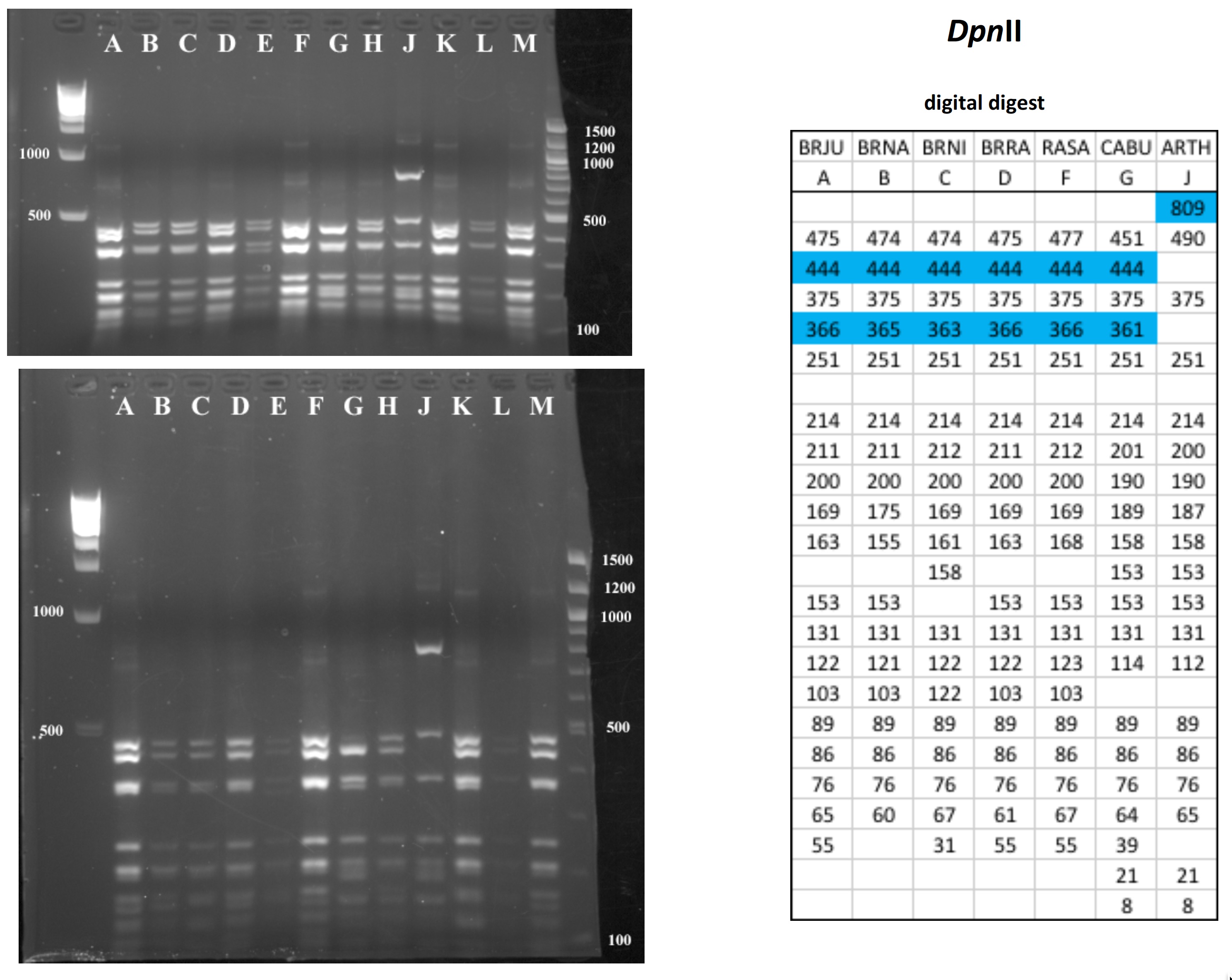
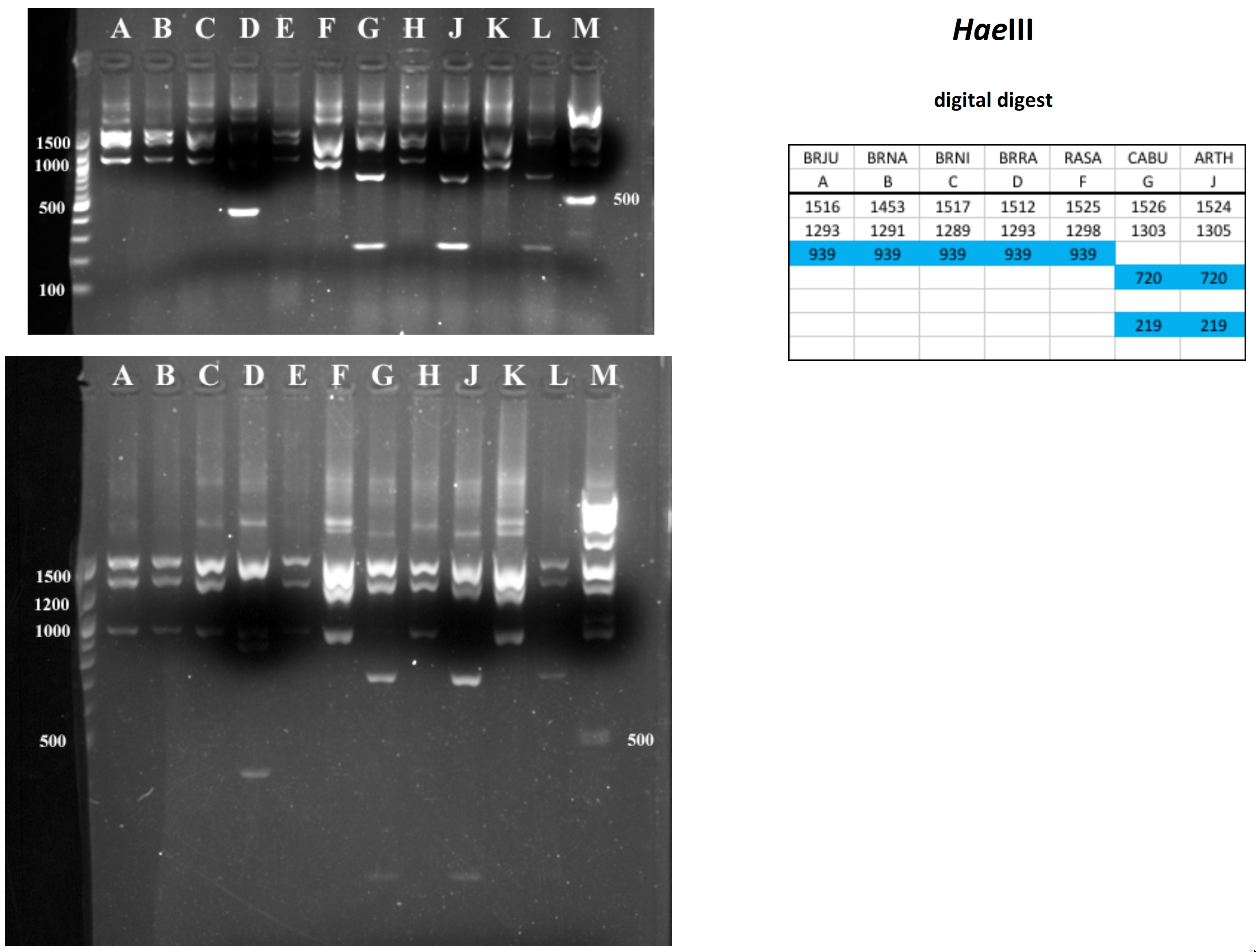
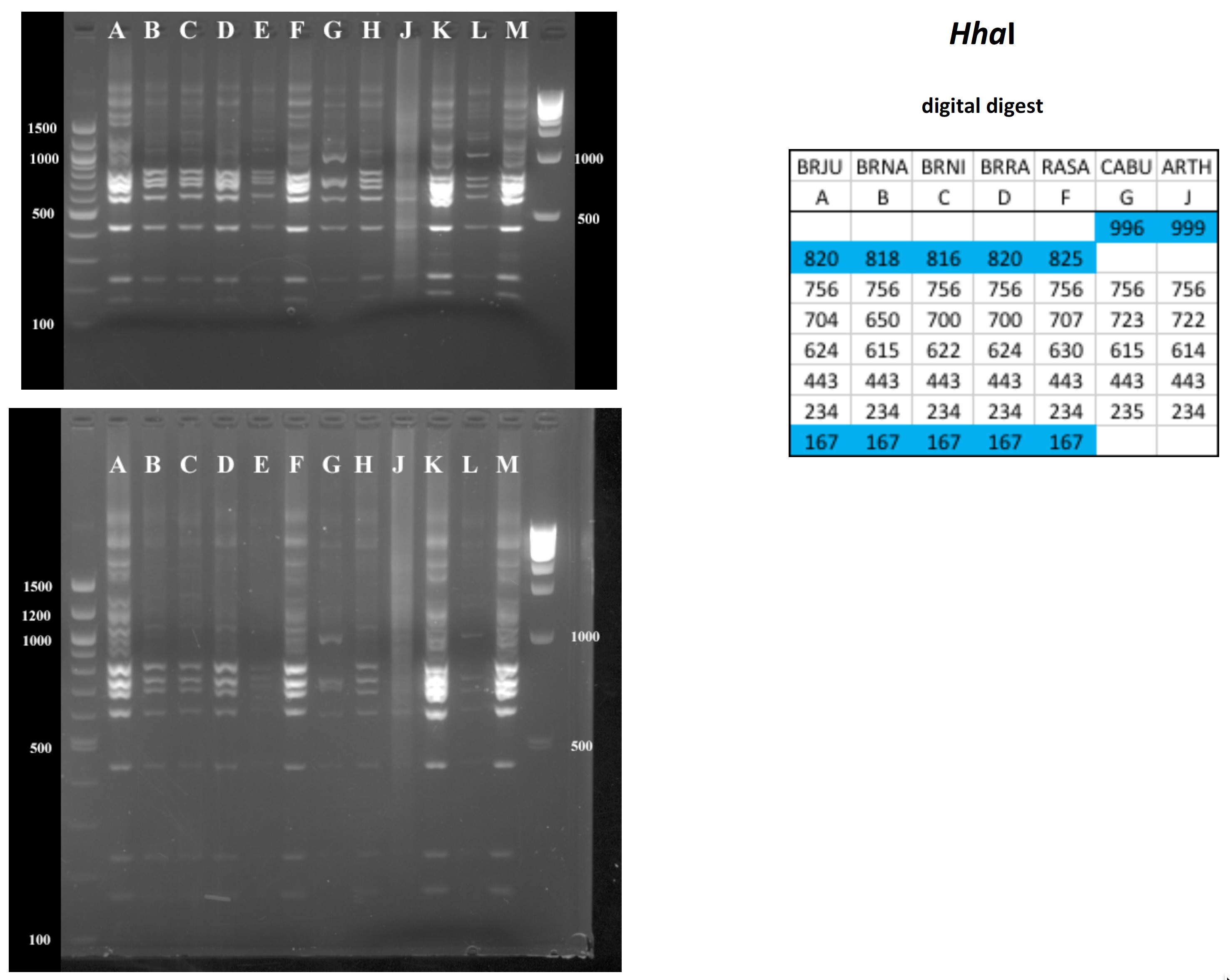
Mustard cpDNA -- 2019 PCR RFLP digests
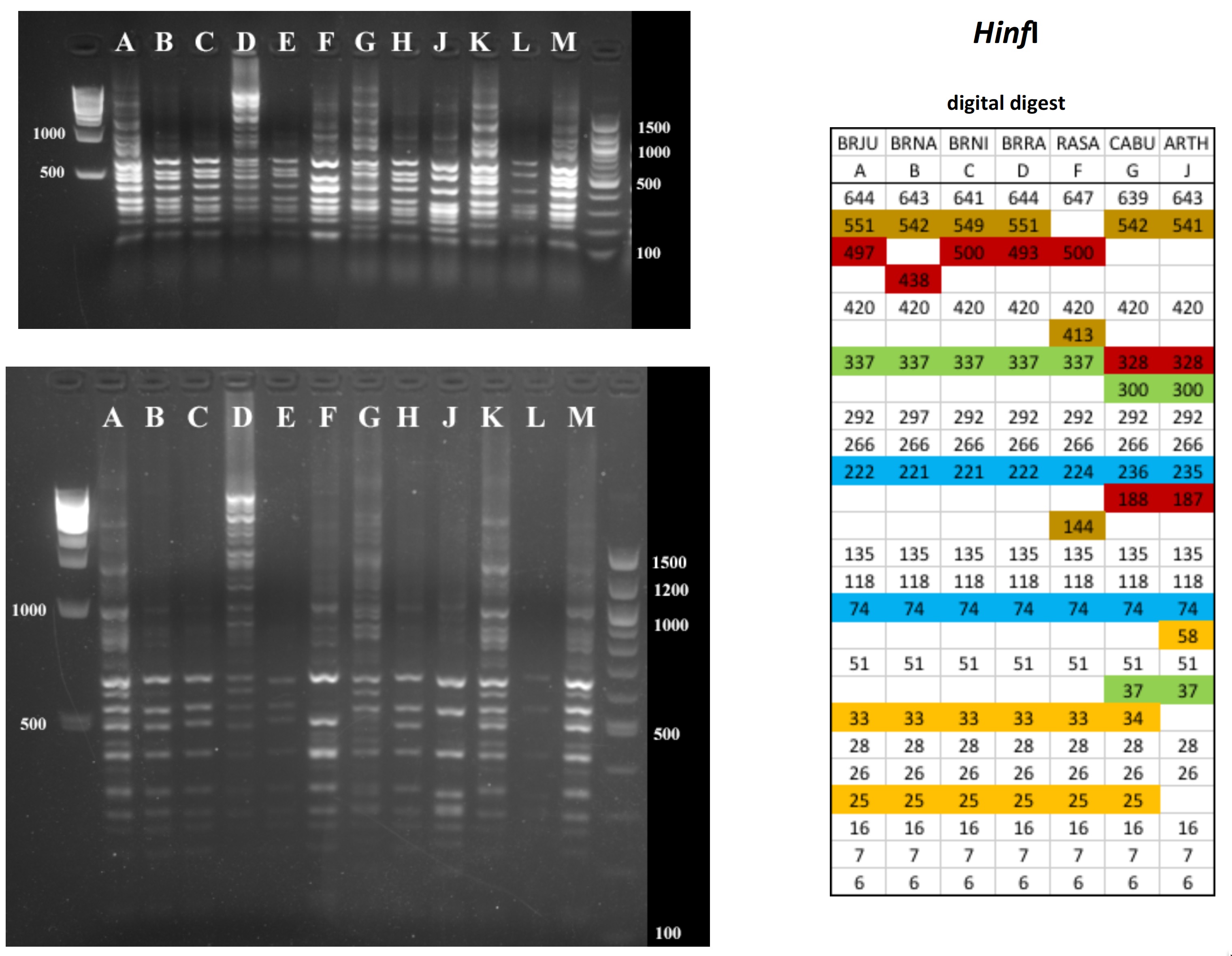
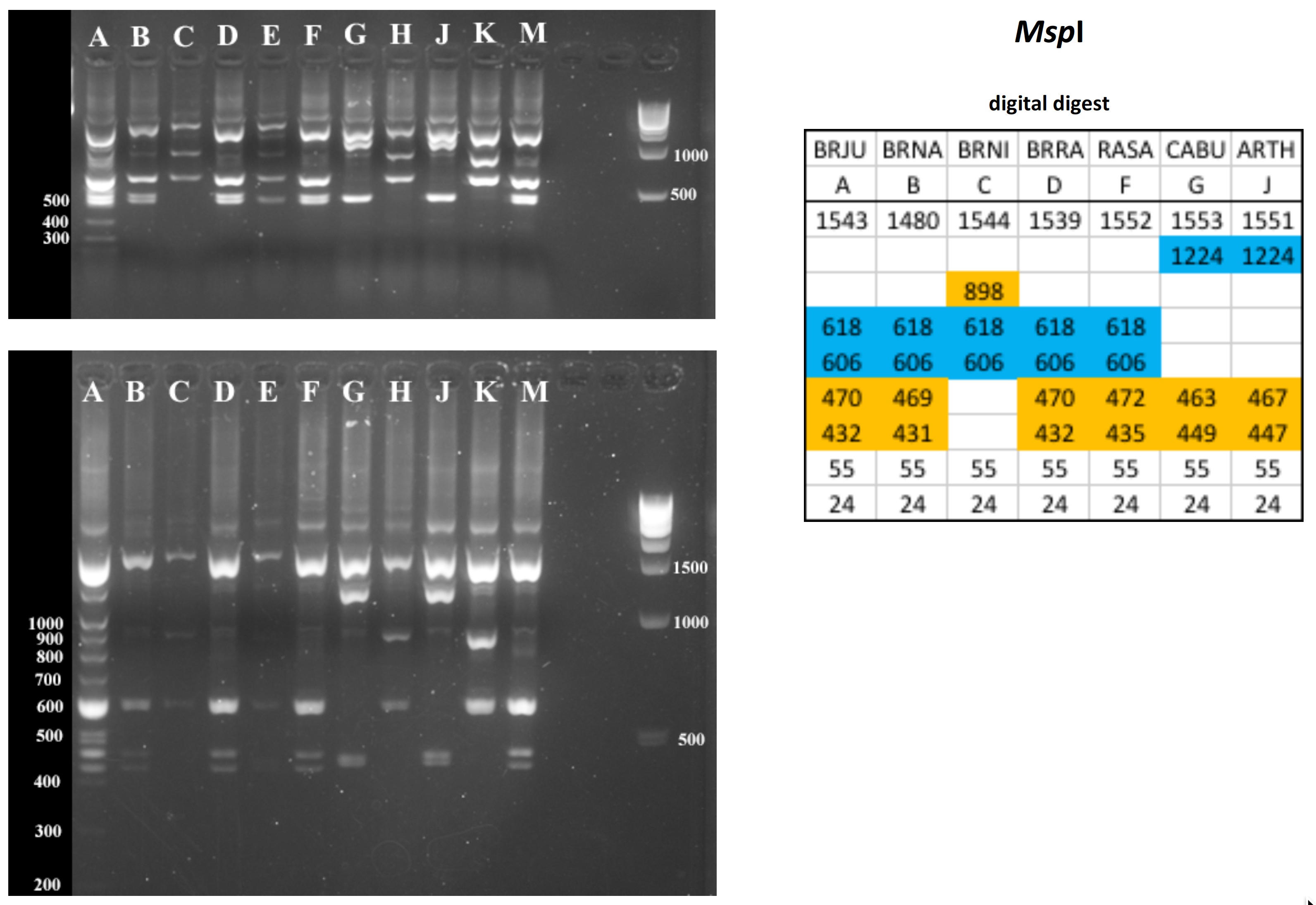
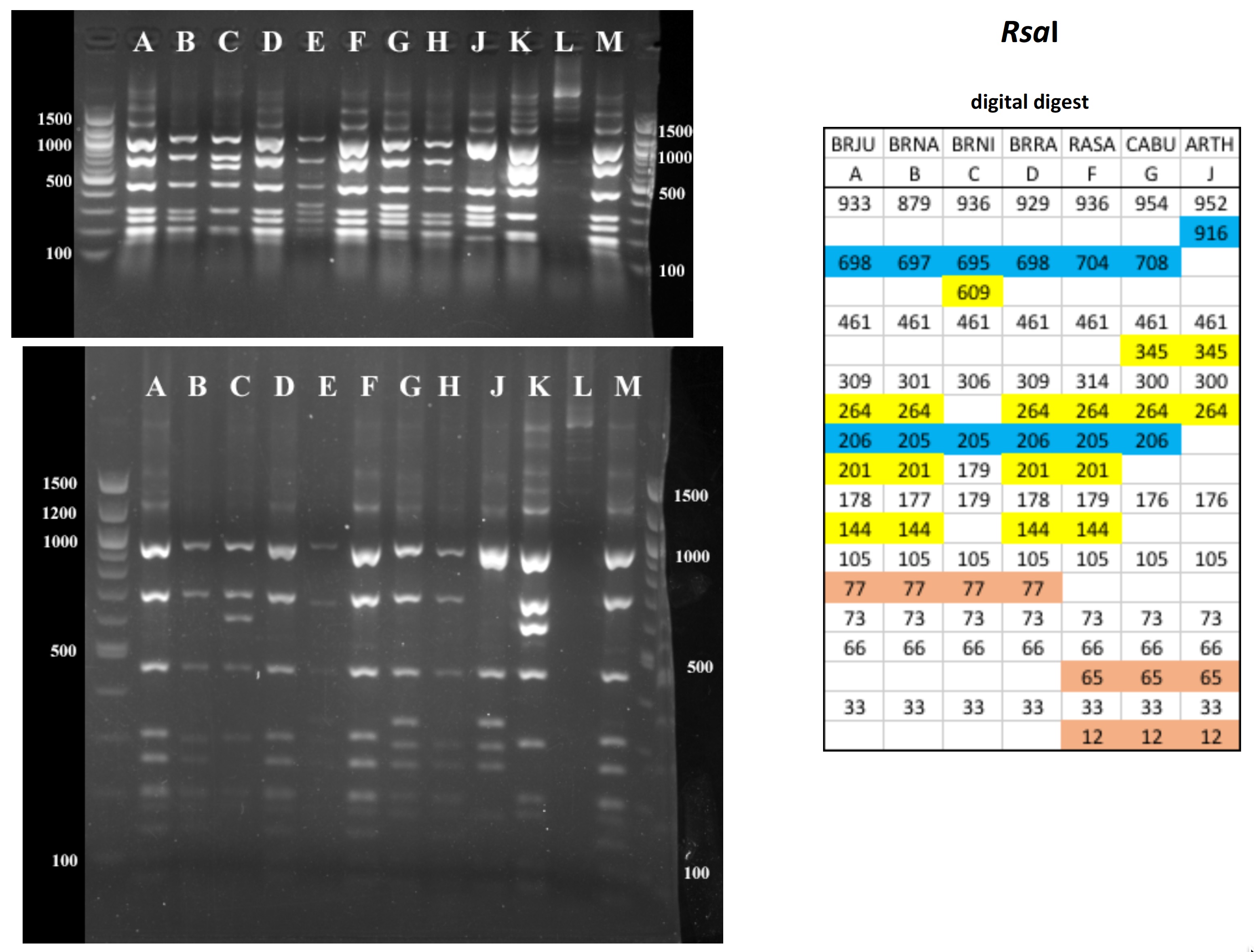
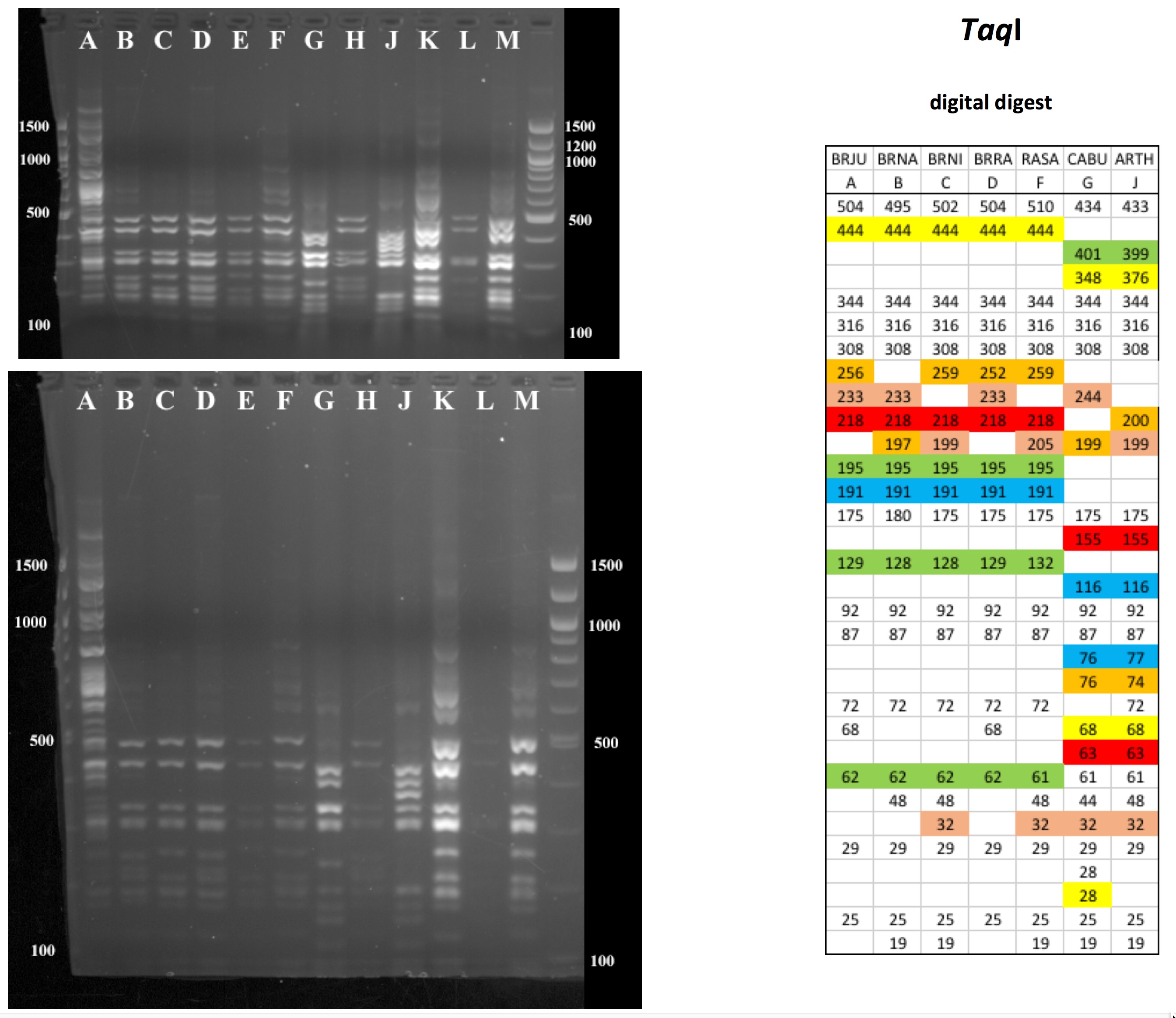
All digests below include ladders in the left and/or right lanes. Samples should be arranged alphabetically from left to right (A toH, J to M).
The ladder lanes flanking the digests included a mix of NEB's 100 bp ladder and 1 kb ladder (see images below).
Clicking on any gel image will bring up a new browser window with a higher resolution image of the digest.
Our results this year can be compared with those from 2004, 2005, 2007, 2009, 2010, 2011, 2013, 2014, 2016, 2017, or 2018.
Digital Digests: Complete chloroplast (cpDNA) genomes were downloaded from GenBank using the 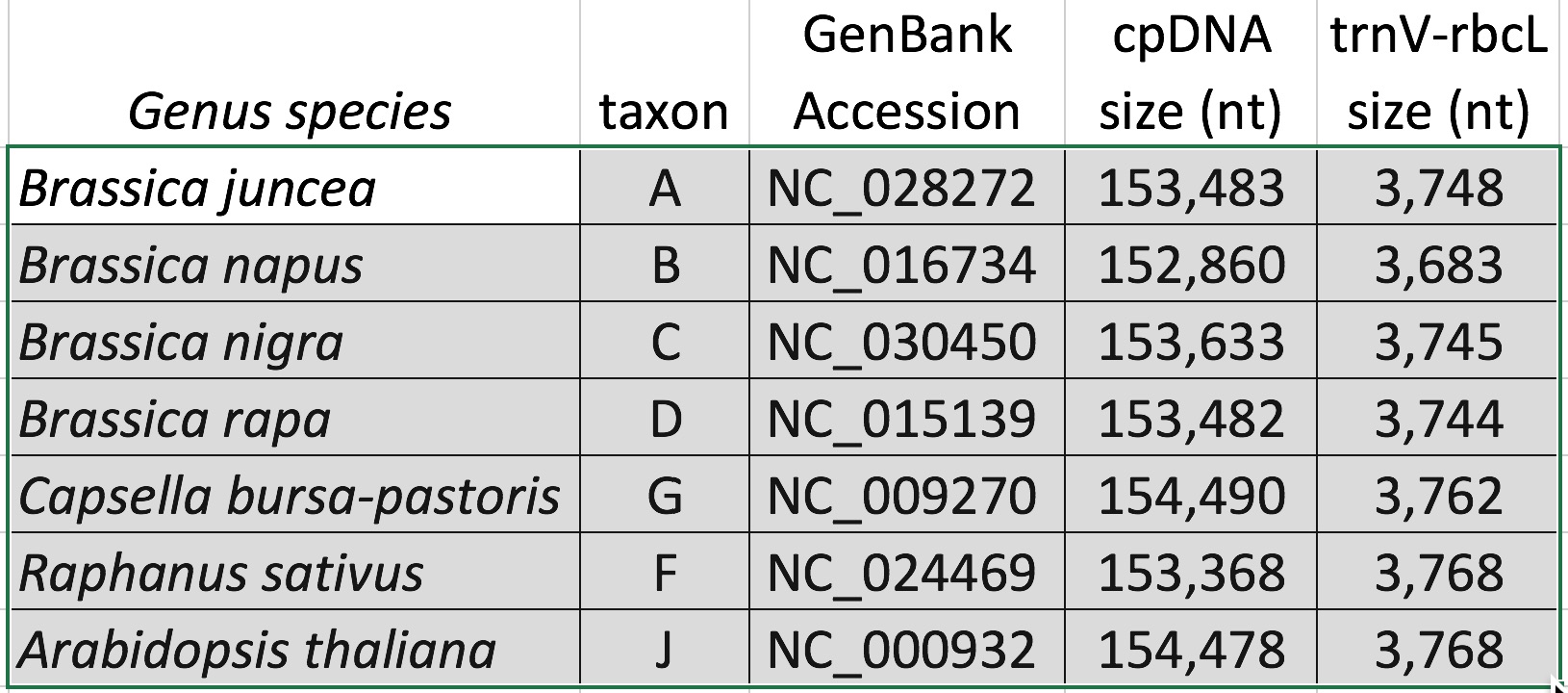 accession numbers in the table to the right. The forward (trnV: CgAACCgTAgACCTTCTCgg) and reverse (rbcL: gCTTTAgTCTCTgTTTgTgg) cpDNA primer sequences were located in each chloroplast genome using Seqeuncher and their position recorded. The complete cpDNA was then digitally digested for each species using the WATCUT online restriction website analysis tool. All cut-sites located between the 5' position of the trnV forward primer and the 5' position of the rbcL reverse primer (on the complimentary strand) were recorded for each species.
accession numbers in the table to the right. The forward (trnV: CgAACCgTAgACCTTCTCgg) and reverse (rbcL: gCTTTAgTCTCTgTTTgTgg) cpDNA primer sequences were located in each chloroplast genome using Seqeuncher and their position recorded. The complete cpDNA was then digitally digested for each species using the WATCUT online restriction website analysis tool. All cut-sites located between the 5' position of the trnV forward primer and the 5' position of the rbcL reverse primer (on the complimentary strand) were recorded for each species.
The sequence-ordered list of expected fragment sizes was then calculated and compared between species to identify homologous cut-sites that were conserved and those that were variable. For cut-sites that varied between taxa, the sequence-ordered fragment list was used to associate i) the two band sizes predicted to result from a cut-site that was present in one or more species with ii) the homologous, uncut band size in the species that lacked the cut-site. The list of cut-site fragments for each species was then reordered by fragment size to provide a predicted digital digest showing the size order bands would be observed on a gel. Bands that represented the cut and uncut homologous fragments were color coded in the tables below for each restriction enzyme to make assessment of the variation easier to visually interpret.
NEB 100bp ladder  |
NEB 1 kbladder 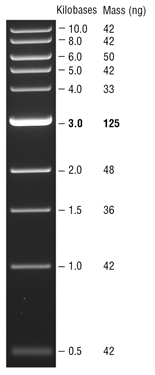 |